Display Models¶
Generate the example file¶
To run the examples of this section, you must first move to the ~/pnbody_examples directory.
If the latter does not exists, you can first create it with:
pNbody_copy-examples
then inside this directory, type:
./scripts/mkmodel_for_display.py
This will create the N-body file snapd.dat.
This file contains a rotating disk of radius 30, with a small plummer sphere centered on (15,15,10).
The display method¶
Any model may be displayed simply using the method NbodyDefault.display().
This method takes several parameters that will be described in detail below.
For our first example, we simply use the size parameter, which set the size of
the displayed area:
>>> from pNbody import *
>>> nb = Nbody("snapd.dat",ftype='gadget')
>>> nb.display(size=(50,50))
If your default parameters are still the ones of the pNbody distribution, this should display the following image:
Display parameters¶
pNbody provides rather complex tools to display N-body systems. The tools uses different parameters that are sumarized in the following table:
name |
meaning |
value |
type |
|---|---|---|---|
obs |
observer |
None |
(ArrayObs) |
xp |
observing position |
None |
(List) |
x0 |
position of observer |
None |
(List) |
alpha |
angle of the head |
None |
(Float) |
view |
view |
xz |
(String) |
r_obs |
dist. to the observer |
201732.223771 |
(Float) |
clip |
clip planes |
(100866.11188556443, 403464.44754225772) |
(Tuple) |
cut |
cut clip planes |
no |
(String) |
eye |
name of the eye |
None |
(String) |
dist_eye |
distance between eyes |
-0.0005 |
(Float) |
foc |
focal |
300.0 |
(Float) |
persp |
perspective |
off |
(String) |
shape |
shape of the image |
(512, 512) |
(Tuple) |
size |
pysical size |
(6000, 6000) |
(Tuple) |
frsp |
frsp |
0.0 |
(Float) |
space |
space |
pos |
(String) |
mode |
mode |
m |
(String) |
rendering |
rendering mode |
map |
(String) |
filter_name |
name of the filter |
None |
(String) |
filter_opts |
filter options |
[10, 10, 2, 2] |
(List) |
scale |
scale |
log |
(String) |
cd |
cd |
0.0 |
(Float) |
mn |
mn |
0.0 |
(Float) |
mx |
mx |
0.0 |
(Float) |
l_n |
number of levels |
15 |
(Int) |
l_min |
min level |
0.0 |
(Float) |
l_max |
max level |
0.0 |
(Float) |
l_kx |
l_kx |
10 |
(Int) |
l_ky |
l_ky |
10 |
(Int) |
l_color |
level color |
0 |
(Int) |
l_crush |
crush background |
no |
(String) |
b_weight |
box line weight |
0 |
(Int) |
b_xopts |
x axis options |
None |
(Tuple) |
b_yopts |
y axis options |
None |
(Tuple) |
b_color |
line color |
255 |
(Int) |
The value of these parameters may be obtained by the command:
pNbody_show-parameters
Set the observer position¶
When creating an image from a model, one has to choose the observer position, the look at point and the orientation of the head. The user has tree possibilities to define these parameters :
Define manually the observer matrix
obs.obsis a 4x3 array matrix. Meaning of the four vectors composing this matrix is given in the following table :
obs
Meaning
obs[0]
position of the observer
obs[1]
position of the look at point (with respect to the position of the observer)
obs[2]
position of the head (with respect to the position of the observer)
obs[3]
position of the arm (with respect to the position of the observer)
If
obsis defined, it is used in priority.
Using the parameters
xp,x0andalpha, wherex0is the observer position,xpis the look at point andalphathe angle between the head and the z axis.Using the parameters
viewandr_obs. This simpler method is used ifobs,xpandx0are set toNone. The parameterviewcan be equal to xy, xz or yz, the projection being parallel to one of the main axis.r_obsgives the distance between the observer and the look at point.
Example:¶
To see the disk face-on, projecting it along the z axis:
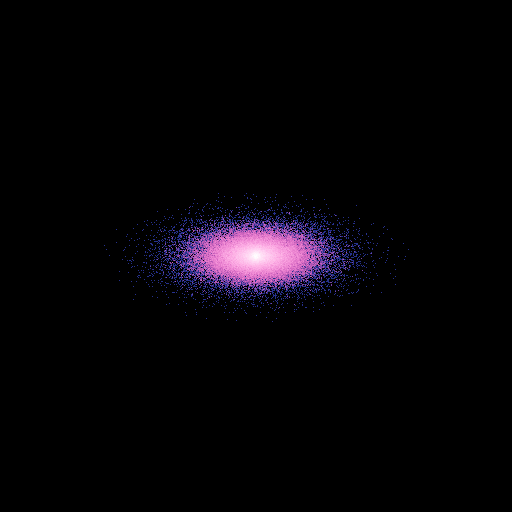
>>> nb.display(obs=None,x0=None,xp=None,size=(50,50),view='xy')

To align the center of the disk with the center of the sphere:
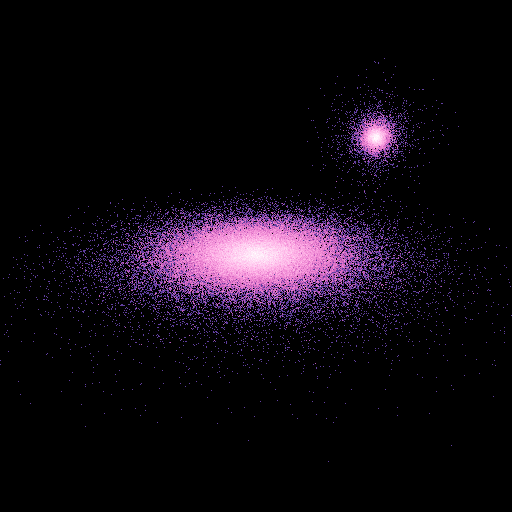
>>> nb.display(obs=None,x0=[30.,30.,20.],xp=[15,15,10],alpha=0,size=(50,50))
To look at the disk from the bottom, tilting the head from 45 degres:
>>> nb.display(obs=None,x0=[0,-50,-50],xp=[0,0,0],alpha=pi/4,size=(50,50))

Set the projection mode¶
pNbody offers two projection modes. If persp``='on', the model is projectec using a frustrum projection matrix.
In the other case, it uses an ortho matrix (orthogonal projection). The near and far clipping planes are given by
the parameter ``clip containing a tuple. The left, right, bottom and top clipping planes are given by the parameter size.
If cut is set to ‘yes’, particles outside the box defined by the 6 planes are not displayed.
Example:¶
Using the frustrum projection:
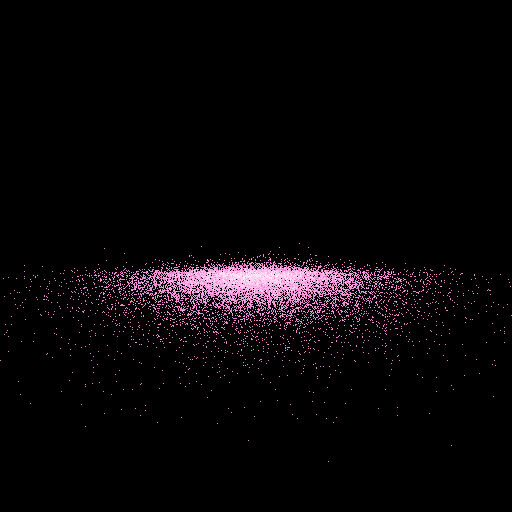
>>> nb.display(obs=None,x0=[0,-50,20],xp=[0,0,0],alpha=0,size=(5,5),persp='on',clip=(10,50))
The field of view is determined using clip and size.
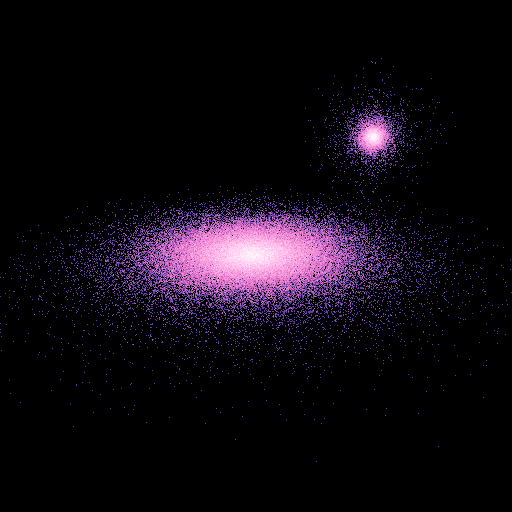
If cut is set to yes, only particles inside the clip planes are displayed:
>>> nb.display(obs=None,x0=[0,-50,20],xp=[0,0,0],alpha=0,size=(5,5),persp='on',clip=(10,50),cut='yes')
Stero mode¶
In order to create stereo images, you can specify the eye you are looking with (‘right’,’left’).
When exposition the model using the method NbodyDefault.expose(), the observer will be rotate around
an axis parallel to its head, with a center rotation in the direction of the look at point, at a distance
foc of the eye. The angle of the rotation is such as to move the observer of a distance dist_eye/2.
Example:¶
Left and right images may be obtained respectively with:
>>> nb.display(obs=None,x0=[0,-50,20],xp=[0,0,0],alpha=0,size=(5,5),persp='on',clip=(10,50),foc=100,dist_eye=0.01,eye='left')
>>> nb.display(obs=None,x0=[0,-50,20],xp=[0,0,0],alpha=0,size=(5,5),persp='on',clip=(10,50),foc=100,dist_eye=0.01,eye='right')
This gives the following image that may be seen using a cross-eyed vision:
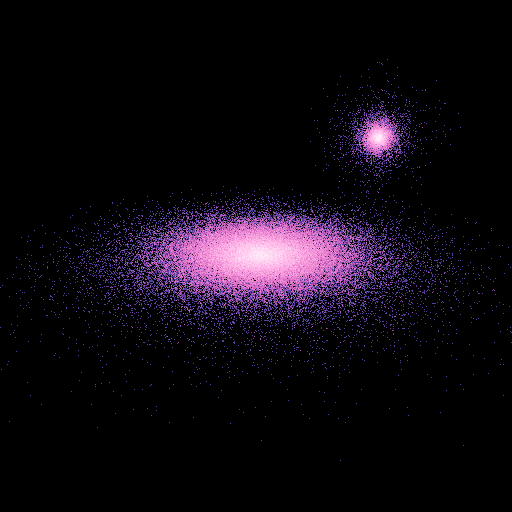
Shape of the final image¶
The shape in pixel of the final image is set by the tuple shape.
Display space¶
The space parameter may be either pos (default) or vel.
In the second case, the model is displayed in velocity space, this simply means
that the positions are replaced by the velocities.
Example:¶
The model in the velocity space gives:
>>> nb.display(obs=None,x0=None,size=(1,1),view='xy',space='vel')
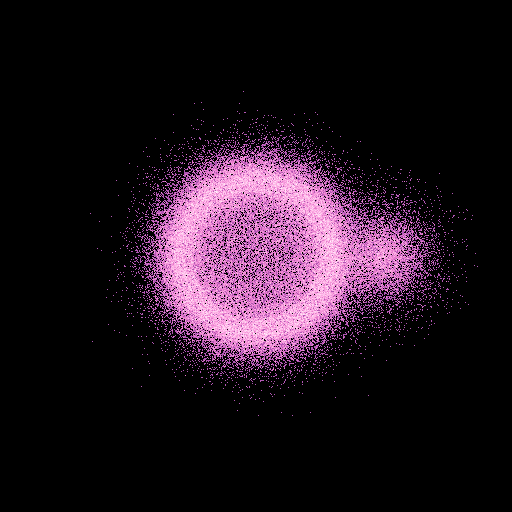
We clearly see the ofset of (0.5,0,0) in velocity of the sphere.
Display mode¶
The mode parameter is very important. It tells the method NbodyDefault.display() which
physical quantities must be displayed. By default, it value is m, meaning the mass. When used,
a projected mass map (surface density) is returned.
The value of mode parameter may be set to:
a value from the following list. In this case, the value does not dependends on the observer position :
Value
Meaning
Formula
“m”
zero momentum
\(\sum m\)
“x”
first momentum in x
“y”
first momentum in y
“z”
first momentum in z
“x2”
second momentum in x
“y2”
second momentum in y
“z2”
second momentum in z
“vx”
first velocity momentum in x
\(\sum m vx\)
“vy”
first velocity momentum in y
\(\sum m vy\)
“vz”
first velocity momentum in z
\(\sum m vz\)
“vx2”
second velocity momentum in x
\(\sum m vx^2\)
“vy2”
second velocity momentum in y
\(\sum m vy^2\)
“vz2”
second velocity momentum in z
\(\sum m vz^2\)
“lx”
first specific kinetic momemtum in x
“ly”
first specific kinetic momemtum in y
“lz”
first specific kinetic momemtum in z
“Lx”
first kinetic momemtum in x
“Ly”
first kinetic momemtum in y
“Lz”
first kinetic momemtum in z
“u”
first momentum of specific energy
“rho”
first momentum of density
“T”
first momentum of temperature
“A”
first momentum of entropy
“P”
first momentum of pressure
“Tcool”
first momentum of cooling time
“Lum”
first momentum of luminosity
“Ne”
first momentum of electronic density
a value from the following list. In this case, the value does dependends on the observer position :
Value
Meaning
Formula
“r”
first momentum of radial distance
“r2”
second momentum of radial distance
“vr”
first momentum of radial velocity
“vr2”
second momemtum of radial velocity
“vxyr”
first momentum of radial velocity in the plane
\(\sum m (x vx + y vy)/\sqrt{x^2+y^2}\)
“vxyr2”
second momentum of radial velocity in the plane
\(\sum m [(x vx + y vy)/\sqrt{x^2+y^2}]^2\)
“vtr”
first momentum of tangential velocity in the plane
\(\sum m (x vx - y vy)/\sqrt{x^2+y^2}\)
“vtr2”
second momentum of tangential velocity in the plane
\(\sum m [(x vx - y vy)/\sqrt{x^2+y^2}]^2\)
any scalar linked with each particle, for example the density
nb.rho.
Example:¶
A radial velocity map is obtained using mode=vr:
>>> nb.display(obs=None,x0=[0,-50,25],xp=[0,0,0],alpha=0,size=(30,30),mode='vr',scale='lin',palette='rainbow4')
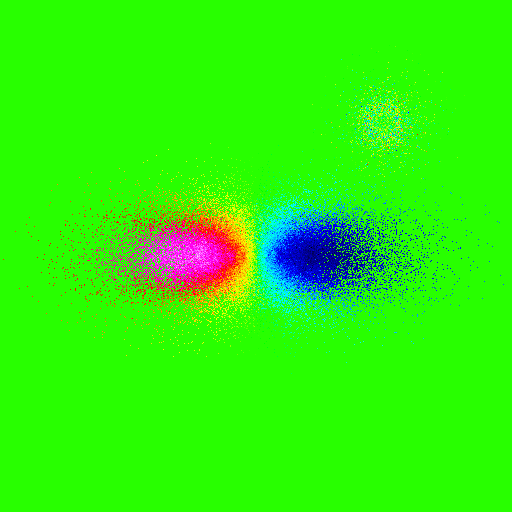
Note that we have used a linear scale here.
Rendering¶
The parameter rendering is by default set to map. This means that particles are projected on a grid (2d histrogram).
However, in some circumstances, it may be usefull to display simple objects, like a cube or a sphere, determined
by a small number of points. The object is obtained by linking all points with segments. This is done by setting
the parameter rendering to one of the following value:
Value
Call
Meaning
lines
draw_lines
draw a continuous line linking all points
segments
draw_segments
draw disconnected segemnts (need persp=’on’)
points
draw_points
draw individual points
polygon
draw_polygon
draw a continuous closed line
polygon2
draw_polygonN 2
draw polygons with groups of 2 points
polygon4
draw_polygonN 4
draw polygons with groups of 4 points
polygon10
draw_polygonN 10
draw polygons with groups of 10 points
polygon#
draw_polygonN n
draw polygons with groups of n points
Examples:¶
>>> from pNbody import ic
>>> nb = ic.box(20,1,1,1)
>>> nb.display(shape=(256,256),size=(1.1,1.1))

>>> nb.display(shape=(256,256),size=(1.1,1.1),rendering='points')

>>> nb.display(shape=(256,256),size=(1.1,1.1),rendering='lines')

>>> nb.display(shape=(256,256),size=(1.1,1.1),rendering='polygon')

>>> nb.display(shape=(256,256),size=(1.1,1.1),rendering='segments',persp='on')
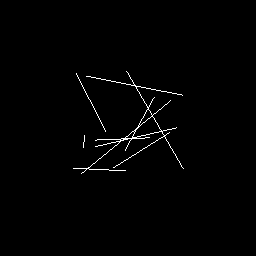
>>> nb.display(shape=(256,256),size=(1.1,1.1),rendering='polygon4')

Lets now display the sphere.dat model
>>> from pNbody import *
>>> nb = Nbody("sphere.dat",ftype='gadget')
>>> nb.display(obs=None,x0=[-50,-50,25],xp=[0,0,0],alpha=0,size=(2,2))
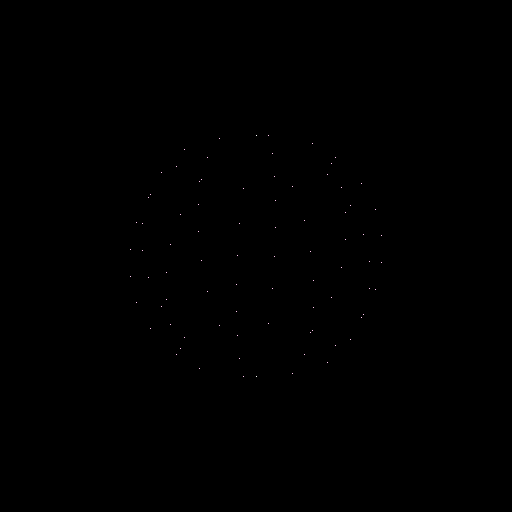
This model is a discretised sphere.
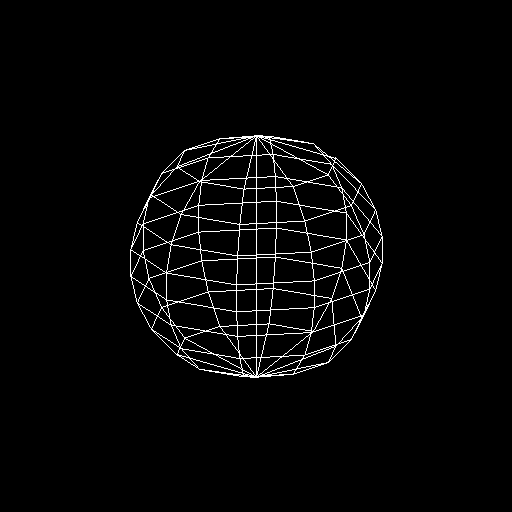
using rendering='polygon' this gives:
>>> nb.display(obs=None,x0=[-50,-50,25],xp=[0,0,0],alpha=0,size=(2,2),rendering='polygon')
Set color range¶
Once a mapping has been performed, it return a matrix containing physical values. The latter must be transformed into an image coded by 256 colors, i.e, a matrix containing integers between 0 and 255. The transformation from the initial matrix and the integer matrix is determined using four parameters:
Parameter
Meaning
scale
the scaling ‘lin’ or ‘log’
cd
if scale=’lin’, this gives the position of the elbow
mn
the minimum physical value
mx
the maximum physical value
In more details, if \(M_p\) is the physical matrix and \(M_i\) the integer one, the transformation is:
when
scale= ‘lin’:
when
scale= ‘log’:
\[M_i = 255 \left( \frac{\log( 1+\frac{M_p-mn}{cd}) }{ \log(1+ \frac{mx-mn}{cd}) } \right)\]
If mn = 0 or mx = 0, or cd = 0, these parameters are set automatically.
Examples:¶
To cut the velocities at a values of -0.3 and 0.3 in the velocity map example:
>>> nb.display(obs=None,x0=[0,-50,25],xp=[0,0,0],alpha=0,size=(30,30),mode='vr',scale='lin',mn=-0.2,mx=0.2,palette='rainbow4')
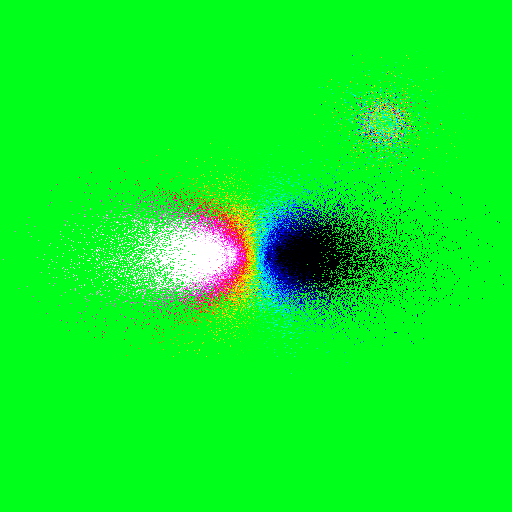
To change the contrast of an image:
>>> nb.display(obs=None,x0=[0,-50,25],xp=[0,0,0],alpha=0,size=(30,30),cd=1e2)

Set filters¶
Its possible to apply filter on the physical matrix, before converting it into an integer matrix.
To set a filter, you must specify the filter_name parameter and the filter_options parameter:
The actual filters are :
Filter name
filter opts
convol
[nx,ny,sx,xy]
convolve
[nx,ny,sx,xy]
boxcar
[nx,ny,sx,xy]
gaussian
[sigma]
uniform
[sigma]
Examples:¶
>>> nb.display(obs=None,x0=[0,-50,25],xp=[0,0,0],alpha=0,size=(30,30),mode='vr',scale='lin',palette='rainbow4',filter_name='gaussian',filter_opts=[5])
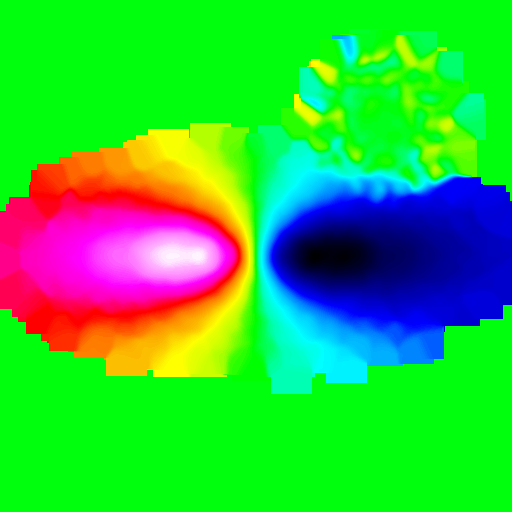
Draw contours¶
It is possible to add contours
Parameter
Meaning
l_n
number of levels
l_min
mininum level value
l_max
maximum level value
l_kx
smoothing size in x
l_ky
smoothing size in x
l_color
color code
l_crush
‘yes’ or ‘no’ , if yes, display only contours
Examples:¶
>>> parameters = {}
>>> parameters['obs'] = None
>>> parameters['x0'] = [0,-50,25]
>>> parameters['xp'] = [0,0,0]
>>> parameters['alpha'] = 0
>>> parameters['size'] = (30,30)
>>> parameters['mode'] = 'vr'
>>> parameters['scale'] = 'lin'
>>> parameters['palette'] = 'rainbow4'
>>> parameters['filter_name'] = 'gaussian'
>>> parameters['filter_opts'] = [5]
>>> parameters['l_n'] = 20
>>> parameters['l_min'] = -0.2
>>> parameters['l_max'] = -0.2
>>> parameters['l_color'] = 255
>>> nb.display(parameters,palette='rainbow4')
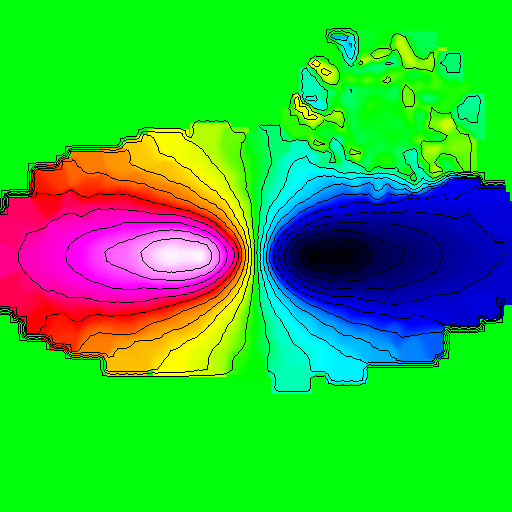
Draw axis¶
It is possible to draw a very simple axis around an image, using the following parameters:
Parameter
Meaning
b_weight
weight of the line
b_xopts
x ticks options (m0,d0,h0,m1,d1,h1) m=dist between ticks, d=first tick h=height of the ticks
b_yopts
y ticks options (m0,d0,h0,m1,d1,h1) m=dist between ticks, d=first tick h=height of the ticks
b_color
smoothing size in x
Examples:¶
>>> parameters['b_weight'] = 1
>>> parameters['b_xopts'] = 1
>>> parameters['b_xopts'] = None
>>> parameters['b_yopts'] = None
>>> parameters['b_color'] = 255
>>> nb.display(parameters,palette='rainbow4')
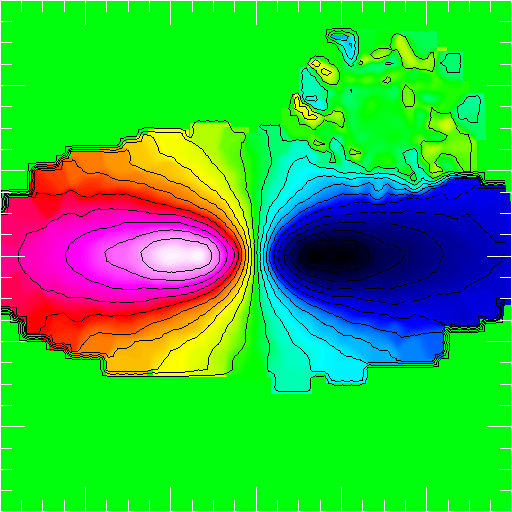
frsp¶
This last parameter is used to smooth the image using an adaptative smoothing length.
Typically, in N-body system, the adaptative smoothing length may be the one
derived using the SPH technique. When the particles are projected in the focal plane,
each particle is convolved whith a kernel corresponding to its proper smoothing length
multiplied by the parameter frsp. This allows to play with the strength of the smooting.
The value of the smoothin length must be stored in th variable nb.rsp..
Examples:¶
Here, we first use the NbodyDefault.ComputeSph() method to compute
the SPH radius of each particle. This radius is determined a the radius
of a sphere containing 50 neighbors with a maximal deviation of 1.
As this method automatically store the results to nb.Hsml we have
to copy it to nb.rsp, before calling the display function:
>>> nb.ComputeSph(DesNumNgb=50,MaxNumNgbDeviation=1)
>>> nb.rsp = nb.Hsml
>>> nb.display(obs=None,x0=None,xp=None,size=(50,50),view='xy',frsp=2)


