Generating mass profiles¶
A serie of routines dedictated to the generation of mass profiles are provided by the ic module.
They can be devided according to their geometry:
function name |
description |
|---|---|
box |
particles in a box |
function name |
description |
|---|---|
homodisk |
homogeneous disk |
kuzmin |
kuzmin disk |
expd |
exponnential disk |
miyamoto_nagai |
Miyamoto-Nagai disk |
function name |
description |
|---|---|
homosphere |
homogeneous sphere |
plummer |
phlummer sphere |
nfw |
Navarro-Frenk-White profile |
hernquist |
Hernquist profile |
dl2 |
\[\rho \sim (1-\epsilon (r/r_{\rm{max}})^2)\]
|
isothm |
Isothermal profile |
pisothm |
Pseudo-Isothermal profile |
shell |
shell |
burkert |
Burkert profile |
nfwg |
Modified Navarro-Frenk-White profile |
generic2c |
generic two slope model |
generic_alpha |
\[\rho \sim (r+e)^a\]
|
generic_Mr |
spherical generic model |
Finally, some routines generate special object used for rendering:
function name |
description |
|---|---|
line |
a simple line |
square |
a square |
circle |
a circle |
grid |
a 2d grid |
cube |
a cube |
sphere |
a sphere |
Each of these routines returns an Nbody object.
Examples:¶
>>> from pNbody import ic >>> nb=ic.box(1000,1,1,1) >>> nb.info() ----------------------------------- particle file : ['box.h5py'] ftype : 'swift' mxntpe : 6 nbody : 1000 nbody_tot : 1000 npart : [1000, 0, 0, 0, 0, 0] npart_tot : [1000, 0, 0, 0, 0, 0] mass_tot : 1.0000001 byteorder : 'little' pio : 'no'len pos : 1000 pos[0] : array([-0.16595599, 0.44064897, -0.99977124], dtype=float32) pos[-1] : array([-0.1209218 , -0.66304606, 0.05817442], dtype=float32) len vel : 1000 vel[0] : array([0., 0., 0.], dtype=float32) vel[-1] : array([0., 0., 0.], dtype=float32) len mass : 1000 mass[0] : 0.001 mass[-1] : 0.001 len num : 1000 num[0] : 0 num[-1] : 999 len tpe : 1000 tpe[0] : 0 tpe[-1] : 0
atime : 0.0 redshift : 0.0 flag_sfr : 0 flag_feedback : 0 flag_cooling : 0 num_files : 1 boxsize : 0.0 omega0 : 0.0 omegalambda : 0.0 hubbleparam : 0.0 flag_age : 0.0 flag_metals : 0.0 nallhw : [0 0 0 0 0 0] critical_energy_spec: 0.0 >>> >>> nb.display()
Generic spherical distributions:¶
In spherical cases, its possible to generate any mass profile, using a
generic function (generic_Mr()). The function takes as argument a vector that
defines the cumulative mass as a function of the radius:
for example, an homogeneous sphere may be obtained by:
>>> from pNbody import ic
>>> import numpy as np
>>> rmax=10
>>> dr=0.01
>>> n = 10000
>>> rs = np.arange(0,rmax,dr)
>>> Mrs = rs**3
>>> nb = ic.generic_Mr(n,rmax=rmax,R=rs,Mr=Mrs,ftype='gadget')
The verctor rs is not necessary linear. This is usefull
to avoid resolution problems, if for example, the mass profile is steep
in some regions. There, intervals between radial bins may be reduced.
In this more funny example, we generate a radial profile corresponding to the following function:
As before, we use the function generic_Mr():
>>> from pNbody import ic
>>> import numpy as np
>>> n = 1000000
>>> rmax = 10.
>>> dr = 0.01
>>> rs = np.arange(0,rmax,dr)
>>> rho = 0.5*(1+np.cos(4*rs) )*np.exp(-rs)
>>> Mrs = np.add.accumulate(rho*rs**2 * dr)
>>> nb = ic.generic_Mr(n,rmax=rmax,R=rs,Mr=Mrs,ftype='gadget')
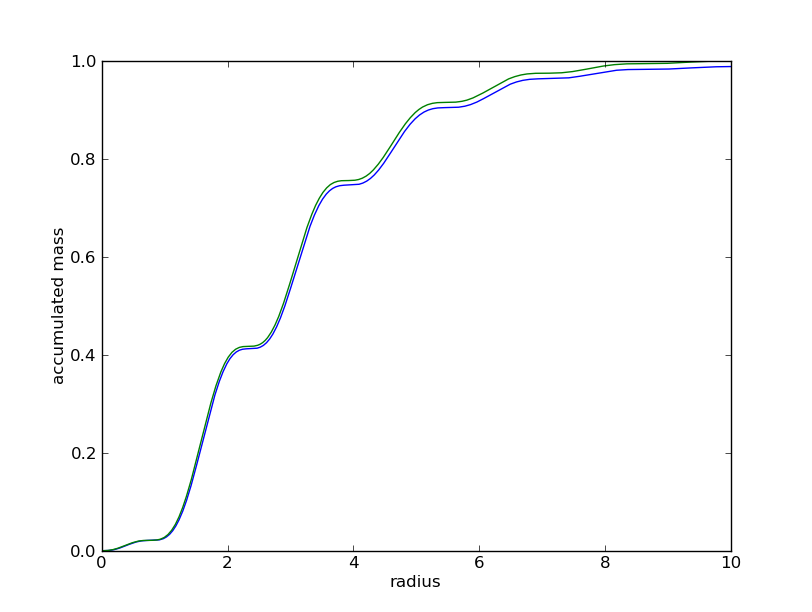
It is possible to check the result by extractig the accumulated mass directely from the Nbody object, using the libgrid module:
>>> from pNbody import libgrid
>>> # create a grid object and extract density and mass profile
>>> G = libgrid.Spherical_1d_Grid(rmin=0,rmax=rmax,nr=512,g=None,gm=None)
>>> r = G.get_r()
>>> rho = G.get_DensityMap(nb)
>>> mr = np.add.accumulate(G.get_MassMap(nb))
>>> # plot results
>>> import pylab as pt
>>> pt.plot(rs,Mrs)
>>> pt.plot(r,mr)
>>> pt.xlabel("radius")
>>> pt.ylabel("accumulated mass")
>>> pt.show()
If everything goes well, we obtain a nice correspondance between the imposed and the computed mass profile, as seen in the graph below.
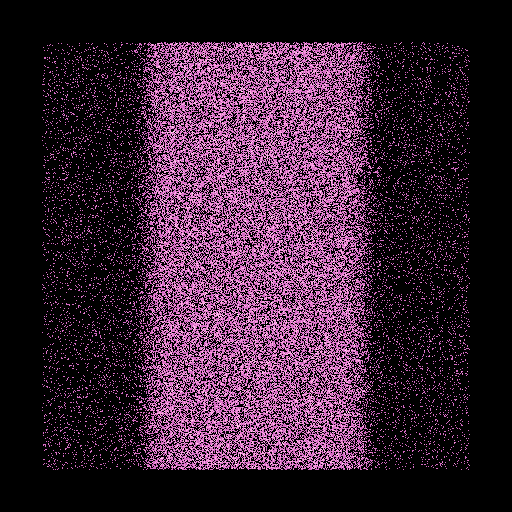
Generic cubic distributions:¶
Its possible generate a cubic box with the density varying along one axix. This is obtained
by using the generic_Mx() function. As for generic_Mr(), we have to define
the variation of the cumulative mass, in this case, the x axis. In the following example,
we set a two level transition along the x axis, following the function:
This gives:
>>> from pNbody import ic
>>> import numpy as np
>>> xs = np.arange(0,1,1./10000.)
>>> n = 100000
>>> rho1 = 1
>>> rho2 = 10
>>> x1 = 0.25
>>> x2 = 0.75
>>> sigma = 0.025
>>> rho = rho1 + (rho2-rho1)/( (1+np.exp(-2.*(xs-x1)/sigma))*(1+np.exp(2.*(xs-x2)/sigma)))
>>> Mxs = np.add.accumulate(rho)
>>> nb = ic.generic_Mx(n,1.0,x=xs,Mx=Mxs,name='tbox.dat',ftype='gadget')
Let’s display now the model:
>>> nb.translate([-0.5,-0.5,-0.5])
>>> nb.show(view='xy',size=(0.6,0.6))
The two density levels are well seen in projection.