pNbody tutorial¶
- official website : http://obswww.unige.ch/~revaz/pNbody/
- tutorial : http://obswww.unige.ch/~revaz/pNbody/rst/Tutorial.html
Using pNbody in the interpreter¶
First, load the python module:
In [1]:
from pNbody import *
Create an empty pNbody object and get info
In [2]:
nb = Nbody()
nb.info()
=================================================================================
TODO WARNING: A function that has not been fully written has been called.
TODO WARNING: This is most likely no reason to worry, but hopefully this message
TODO WARNING: will annoy the developers enough to start clean their code up.
TODO WARNING: Meanwhile, you can just carry on doing whatever you were up to.
=================================================================================
-----------------------------------
particle file : ['file.dat']
ftype : 'default'
mxntpe : 6
nbody : 0
nbody_tot : 0
npart : [0, 0, 0, 0, 0, 0]
npart_tot : [0, 0, 0, 0, 0, 0]
mass_tot : 0.0
byteorder : 'little'
pio : 'no'
Create a pNbody object from positions:
In [4]:
pos = np.ones((10,3),np.float32)
nb = Nbody(pos=pos)
nb.info()
=================================================================================
TODO WARNING: A function that has not been fully written has been called.
TODO WARNING: This is most likely no reason to worry, but hopefully this message
TODO WARNING: will annoy the developers enough to start clean their code up.
TODO WARNING: Meanwhile, you can just carry on doing whatever you were up to.
=================================================================================
-----------------------------------
particle file : ['file.dat']
ftype : 'default'
mxntpe : 6
nbody : 10
nbody_tot : 10
npart : [10, 0, 0, 0, 0, 0]
npart_tot : [10, 0, 0, 0, 0, 0]
mass_tot : 1.0
byteorder : 'little'
pio : 'no'
len pos : 10
pos[0] : array([1., 1., 1.], dtype=float32)
pos[-1] : array([1., 1., 1.], dtype=float32)
len vel : 10
vel[0] : array([0., 0., 0.], dtype=float32)
vel[-1] : array([0., 0., 0.], dtype=float32)
len mass : 10
mass[0] : 0.1
mass[-1] : 0.1
len num : 10
num[0] : 0
num[-1] : 9
len tpe : 10
tpe[0] : 0
tpe[-1] : 0
Create a pNbody object using the ic module:
In [6]:
from pNbody import ic
nb = ic.plummer(10000,1,1,1,1,rmax=20.,M=1,name='plummer.dat',ftype='gadget')
comovingintegration = False
nb.write() # writes the nBody object to file 'plummer.dat' with the 'gadget' format (as specified in the Nbody object)
Read a pNbody from a file¶
In [8]:
nb = Nbody('plummer.dat',ftype='gadget')
The format is gadget
In [9]:
comovingintegration = False
In [10]:
nb.info()
-----------------------------------
particle file : ['plummer.dat']
ftype : 'gadget'
mxntpe : 6
nbody : 10000
nbody_tot : 10000
npart : [10000, 0, 0, 0, 0, 0]
npart_tot : [10000, 0, 0, 0, 0, 0]
mass_tot : 0.99999976
byteorder : 'little'
pio : 'no'
len pos : 10000
pos[0] : array([ 0.08620726, -0.8825551 , -0.68550044], dtype=float32)
pos[-1] : array([-0.6835583 , 2.1199563 , -0.13416442], dtype=float32)
len vel : 10000
vel[0] : array([0., 0., 0.], dtype=float32)
vel[-1] : array([0., 0., 0.], dtype=float32)
len mass : 10000
mass[0] : 1e-04
mass[-1] : 1e-04
len num : 10000
num[0] : 0
num[-1] : 9999
len tpe : 10000
tpe[0] : 0
tpe[-1] : 0
atime : 0.0
redshift : 0.0
flag_sfr : 0
flag_feedback : 0
nall : [10000, 0, 0, 0, 0, 0]
flag_cooling : 0
num_files : 1
boxsize : 0.0
omega0 : 0.0
omegalambda : 0.0
hubbleparam : 0.0
flag_age : 0
flag_metals : 0
nallhw : [0, 0, 0, 0, 0, 0]
flag_entr_ic : 0
critical_energy_spec: 0.0
Get particle properties¶
Particle properties are numpy arrays of size nb.nbody:
In [11]:
nb.pos
Out[11]:
array([[ 0.08620726, -0.8825551 , -0.68550044],
[-0.7158597 , 1.8705164 , -0.16716339],
[-0.0029057 , -0.03153811, -0.03678056],
...,
[ 0.40189224, 0.11727811, 0.06398547],
[-0.06060761, -0.17355472, -0.5085909 ],
[-0.6835583 , 2.1199563 , -0.13416442]], dtype=float32)
In [12]:
nb.vel
Out[12]:
array([[0., 0., 0.],
[0., 0., 0.],
[0., 0., 0.],
...,
[0., 0., 0.],
[0., 0., 0.],
[0., 0., 0.]], dtype=float32)
In [13]:
nb.mass
Out[13]:
array([1.e-04, 1.e-04, 1.e-04, ..., 1.e-04, 1.e-04, 1.e-04], dtype=float32)
In [14]:
nb.num
Out[14]:
array([ 0, 1, 2, ..., 9997, 9998, 9999])
In [15]:
nb.tpe
Out[15]:
array([0, 0, 0, ..., 0, 0, 0])
Get the list of particle properties:
In [16]:
nb.get_list_of_array()
Out[16]:
['mass', 'num', 'pos', 'tpe', 'vel']
Display a model¶
# these two commands have been formatted differently so the website of the tutorial can be generated. # just copy them into a fresh interpreter cell and run them to see what they do. nb.show(size=(10,10)) nb.show(size=(20,20),shape=(256,256),palette='rainbow4')
In [17]:
Get physical values¶
positins x coordinates:
In [18]:
nb.x()
Out[18]:
array([ 0.08620726, -0.7158597 , -0.0029057 , ..., 0.40189224,
-0.06060761, -0.6835583 ], dtype=float32)
center of mass
In [19]:
nb.cm()
Out[19]:
array([ 0.0013582 , -0.04152351, 0.01155748])
angular momentum:
In [20]:
nb.Ltot()
Out[20]:
array([0., 0., 0.], dtype=float32)
Compute potential and acceleration:
In [21]:
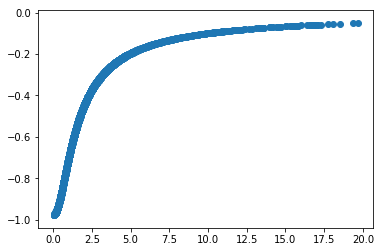
r = nb.rxyz()
pot = nb.Pot(nb.pos,eps=0.1)
acc = nb.Accel(nb.pos,eps=0.1)
alternatively, you can use a treecode algorithm to compute the potential and acceleration (which is much faster):
In [22]:
pot = nb.TreePot(nb.pos,eps=0.1)
acc = nb.TreeAccel(nb.pos,eps=0.1)
create the tree : ErrTolTheta= 0.8
Plot:
In [24]:
import pylab as pt
pt.scatter(r, pot)
pt.show(); # for some reason the plot doesn't show up unless you put the semicolon here.
In [ ]:
