Scipy (Python3) tutorial¶
- official website : http://www.scipy.org/
- documentation : https://docs.scipy.org/doc/scipy/reference/
- Tutorial : https://docs.scipy.org/doc/scipy/reference/tutorial/index.html
Packages¶
| Topic | package name |
|---|---|
| Special functions | scipy.special |
| Integration | scipy.integrate |
| Optimization | scipy.optimize |
| Interpolation | scipy.interpolate |
| Fourier Transforms | scipy.fftpack |
| Signal Processing | scipy.signal |
| Linear Algebra | scipy.linalg |
| Sparse Eigenvalue Problems with ARPACK | scipy.sparse.linalg |
| Compressed Sparse Graph Routines | scipy.sparse.csgraph |
| Spatial data structures and algorithms | scipy.spatial |
| Statistics | scipy.stats |
| Multidimensional image processing | scipy.ndimage |
| File IO | scipy.io |
Integration¶
Load modules:
In [1]:
import numpy as np
from scipy import integrate
help:
In [2]:
help(integrate.quad)
Help on function quad in module scipy.integrate.quadpack:
quad(func, a, b, args=(), full_output=0, epsabs=1.49e-08, epsrel=1.49e-08, limit=50, points=None, weight=None, wvar=None, wopts=None, maxp1=50, limlst=50)
Compute a definite integral.
Integrate func from `a` to `b` (possibly infinite interval) using a
technique from the Fortran library QUADPACK.
Parameters
----------
func : {function, scipy.LowLevelCallable}
A Python function or method to integrate. If `func` takes many
arguments, it is integrated along the axis corresponding to the
first argument.
If the user desires improved integration performance, then `f` may
be a `scipy.LowLevelCallable` with one of the signatures::
double func(double x)
double func(double x, void *user_data)
double func(int n, double *xx)
double func(int n, double *xx, void *user_data)
The ``user_data`` is the data contained in the `scipy.LowLevelCallable`.
In the call forms with ``xx``, ``n`` is the length of the ``xx``
array which contains ``xx[0] == x`` and the rest of the items are
numbers contained in the ``args`` argument of quad.
In addition, certain ctypes call signatures are supported for
backward compatibility, but those should not be used in new code.
a : float
Lower limit of integration (use -numpy.inf for -infinity).
b : float
Upper limit of integration (use numpy.inf for +infinity).
args : tuple, optional
Extra arguments to pass to `func`.
full_output : int, optional
Non-zero to return a dictionary of integration information.
If non-zero, warning messages are also suppressed and the
message is appended to the output tuple.
Returns
-------
y : float
The integral of func from `a` to `b`.
abserr : float
An estimate of the absolute error in the result.
infodict : dict
A dictionary containing additional information.
Run scipy.integrate.quad_explain() for more information.
message
A convergence message.
explain
Appended only with 'cos' or 'sin' weighting and infinite
integration limits, it contains an explanation of the codes in
infodict['ierlst']
Other Parameters
----------------
epsabs : float or int, optional
Absolute error tolerance.
epsrel : float or int, optional
Relative error tolerance.
limit : float or int, optional
An upper bound on the number of subintervals used in the adaptive
algorithm.
points : (sequence of floats,ints), optional
A sequence of break points in the bounded integration interval
where local difficulties of the integrand may occur (e.g.,
singularities, discontinuities). The sequence does not have
to be sorted.
weight : float or int, optional
String indicating weighting function. Full explanation for this
and the remaining arguments can be found below.
wvar : optional
Variables for use with weighting functions.
wopts : optional
Optional input for reusing Chebyshev moments.
maxp1 : float or int, optional
An upper bound on the number of Chebyshev moments.
limlst : int, optional
Upper bound on the number of cycles (>=3) for use with a sinusoidal
weighting and an infinite end-point.
See Also
--------
dblquad : double integral
tplquad : triple integral
nquad : n-dimensional integrals (uses `quad` recursively)
fixed_quad : fixed-order Gaussian quadrature
quadrature : adaptive Gaussian quadrature
odeint : ODE integrator
ode : ODE integrator
simps : integrator for sampled data
romb : integrator for sampled data
scipy.special : for coefficients and roots of orthogonal polynomials
Notes
-----
**Extra information for quad() inputs and outputs**
If full_output is non-zero, then the third output argument
(infodict) is a dictionary with entries as tabulated below. For
infinite limits, the range is transformed to (0,1) and the
optional outputs are given with respect to this transformed range.
Let M be the input argument limit and let K be infodict['last'].
The entries are:
'neval'
The number of function evaluations.
'last'
The number, K, of subintervals produced in the subdivision process.
'alist'
A rank-1 array of length M, the first K elements of which are the
left end points of the subintervals in the partition of the
integration range.
'blist'
A rank-1 array of length M, the first K elements of which are the
right end points of the subintervals.
'rlist'
A rank-1 array of length M, the first K elements of which are the
integral approximations on the subintervals.
'elist'
A rank-1 array of length M, the first K elements of which are the
moduli of the absolute error estimates on the subintervals.
'iord'
A rank-1 integer array of length M, the first L elements of
which are pointers to the error estimates over the subintervals
with ``L=K`` if ``K<=M/2+2`` or ``L=M+1-K`` otherwise. Let I be the
sequence ``infodict['iord']`` and let E be the sequence
``infodict['elist']``. Then ``E[I[1]], ..., E[I[L]]`` forms a
decreasing sequence.
If the input argument points is provided (i.e. it is not None),
the following additional outputs are placed in the output
dictionary. Assume the points sequence is of length P.
'pts'
A rank-1 array of length P+2 containing the integration limits
and the break points of the intervals in ascending order.
This is an array giving the subintervals over which integration
will occur.
'level'
A rank-1 integer array of length M (=limit), containing the
subdivision levels of the subintervals, i.e., if (aa,bb) is a
subinterval of ``(pts[1], pts[2])`` where ``pts[0]`` and ``pts[2]``
are adjacent elements of ``infodict['pts']``, then (aa,bb) has level l
if ``|bb-aa| = |pts[2]-pts[1]| * 2**(-l)``.
'ndin'
A rank-1 integer array of length P+2. After the first integration
over the intervals (pts[1], pts[2]), the error estimates over some
of the intervals may have been increased artificially in order to
put their subdivision forward. This array has ones in slots
corresponding to the subintervals for which this happens.
**Weighting the integrand**
The input variables, *weight* and *wvar*, are used to weight the
integrand by a select list of functions. Different integration
methods are used to compute the integral with these weighting
functions. The possible values of weight and the corresponding
weighting functions are.
========== =================================== =====================
``weight`` Weight function used ``wvar``
========== =================================== =====================
'cos' cos(w*x) wvar = w
'sin' sin(w*x) wvar = w
'alg' g(x) = ((x-a)**alpha)*((b-x)**beta) wvar = (alpha, beta)
'alg-loga' g(x)*log(x-a) wvar = (alpha, beta)
'alg-logb' g(x)*log(b-x) wvar = (alpha, beta)
'alg-log' g(x)*log(x-a)*log(b-x) wvar = (alpha, beta)
'cauchy' 1/(x-c) wvar = c
========== =================================== =====================
wvar holds the parameter w, (alpha, beta), or c depending on the weight
selected. In these expressions, a and b are the integration limits.
For the 'cos' and 'sin' weighting, additional inputs and outputs are
available.
For finite integration limits, the integration is performed using a
Clenshaw-Curtis method which uses Chebyshev moments. For repeated
calculations, these moments are saved in the output dictionary:
'momcom'
The maximum level of Chebyshev moments that have been computed,
i.e., if ``M_c`` is ``infodict['momcom']`` then the moments have been
computed for intervals of length ``|b-a| * 2**(-l)``,
``l=0,1,...,M_c``.
'nnlog'
A rank-1 integer array of length M(=limit), containing the
subdivision levels of the subintervals, i.e., an element of this
array is equal to l if the corresponding subinterval is
``|b-a|* 2**(-l)``.
'chebmo'
A rank-2 array of shape (25, maxp1) containing the computed
Chebyshev moments. These can be passed on to an integration
over the same interval by passing this array as the second
element of the sequence wopts and passing infodict['momcom'] as
the first element.
If one of the integration limits is infinite, then a Fourier integral is
computed (assuming w neq 0). If full_output is 1 and a numerical error
is encountered, besides the error message attached to the output tuple,
a dictionary is also appended to the output tuple which translates the
error codes in the array ``info['ierlst']`` to English messages. The
output information dictionary contains the following entries instead of
'last', 'alist', 'blist', 'rlist', and 'elist':
'lst'
The number of subintervals needed for the integration (call it ``K_f``).
'rslst'
A rank-1 array of length M_f=limlst, whose first ``K_f`` elements
contain the integral contribution over the interval
``(a+(k-1)c, a+kc)`` where ``c = (2*floor(|w|) + 1) * pi / |w|``
and ``k=1,2,...,K_f``.
'erlst'
A rank-1 array of length ``M_f`` containing the error estimate
corresponding to the interval in the same position in
``infodict['rslist']``.
'ierlst'
A rank-1 integer array of length ``M_f`` containing an error flag
corresponding to the interval in the same position in
``infodict['rslist']``. See the explanation dictionary (last entry
in the output tuple) for the meaning of the codes.
Examples
--------
Calculate :math:`\int^4_0 x^2 dx` and compare with an analytic result
>>> from scipy import integrate
>>> x2 = lambda x: x**2
>>> integrate.quad(x2, 0, 4)
(21.333333333333332, 2.3684757858670003e-13)
>>> print(4**3 / 3.) # analytical result
21.3333333333
Calculate :math:`\int^\infty_0 e^{-x} dx`
>>> invexp = lambda x: np.exp(-x)
>>> integrate.quad(invexp, 0, np.inf)
(1.0, 5.842605999138044e-11)
>>> f = lambda x,a : a*x
>>> y, err = integrate.quad(f, 0, 1, args=(1,))
>>> y
0.5
>>> y, err = integrate.quad(f, 0, 1, args=(3,))
>>> y
1.5
Calculate :math:`\int^1_0 x^2 + y^2 dx` with ctypes, holding
y parameter as 1::
testlib.c =>
double func(int n, double args[n]){
return args[0]*args[0] + args[1]*args[1];}
compile to library testlib.*
::
from scipy import integrate
import ctypes
lib = ctypes.CDLL('/home/.../testlib.*') #use absolute path
lib.func.restype = ctypes.c_double
lib.func.argtypes = (ctypes.c_int,ctypes.c_double)
integrate.quad(lib.func,0,1,(1))
#(1.3333333333333333, 1.4802973661668752e-14)
print((1.0**3/3.0 + 1.0) - (0.0**3/3.0 + 0.0)) #Analytic result
# 1.3333333333333333
Methods for Integrating Functions given function object:¶
In [3]:
integrate.quad(np.cos,0,np.pi/2.)
Out[3]:
(0.9999999999999999, 1.1102230246251564e-14)
Methods for Integrating Functions given fixed samples:¶
In [4]:
x = np.arange(0,np.pi/2.,np.pi/100.)
y = np.cos(x)
integrate using the trapeze method:
In [5]:
integrate.trapz(y,x)
Out[5]:
0.99942435289390408
integrate using the simpson method:
In [6]:
integrate.simps(y,x)
Out[6]:
0.99950521309271656
Optimization¶
Load module:
In [7]:
from scipy.optimize import leastsq
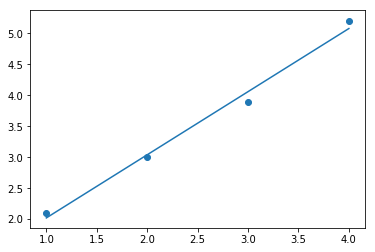
Fit a linear regression to data¶
start wiith some data:
In [8]:
x = np.array([1,2,3,4],float)
y = np.array([2.1,3,3.9,5.2],float)
define a fitting function:
In [9]:
def linfct(x,a,b):
return a*x + b
define a residual function
In [10]:
def residuals(p, x, y):
a,b = p
err = y-linfct(x,a,b)
return err
first guess
In [11]:
p0 = 1.,1.
find optimal values
In [12]:
plsq,cmt = leastsq(residuals, p0, args=(x, y))
a = plsq[0]
b = plsq[1]
a,b
Out[12]:
(1.0200000000000435, 1.0)
compute the fit
In [13]:
yf = linfct(x,a,b)
now, plot:
In [14]:
import pylab as plt
plt.scatter(x,y)
plt.plot(x,yf)
plt.show()
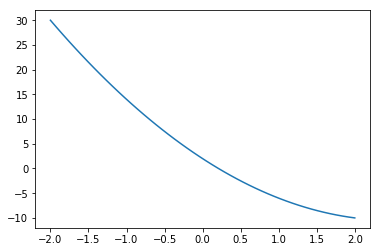
Root finding routines¶
Define a function
In [15]:
def f(x):
return 2*x*x - 10*x + 2
In [16]:
import pylab as plt
x = np.linspace(-2,2,50)
plt.plot(x,f(x))
plt.show()
Using the Newtown method to solve 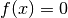
In [17]:
from scipy.optimize import newton
x0 = 0
newton(f,x0)
Out[17]:
0.20871215252207997
Using the Bissection method
In [18]:
from scipy.optimize import bisect
xmin=-2
xmax=2
bisect(f,xmin,xmax)
Out[18]:
0.20871215252373077
Interpolation¶
Load module
In [19]:
from scipy import interpolate
In [20]:
help(interpolate.splrep)
Help on function splrep in module scipy.interpolate.fitpack:
splrep(x, y, w=None, xb=None, xe=None, k=3, task=0, s=None, t=None, full_output=0, per=0, quiet=1)
Find the B-spline representation of 1-D curve.
Given the set of data points ``(x[i], y[i])`` determine a smooth spline
approximation of degree k on the interval ``xb <= x <= xe``.
Parameters
----------
x, y : array_like
The data points defining a curve y = f(x).
w : array_like, optional
Strictly positive rank-1 array of weights the same length as x and y.
The weights are used in computing the weighted least-squares spline
fit. If the errors in the y values have standard-deviation given by the
vector d, then w should be 1/d. Default is ones(len(x)).
xb, xe : float, optional
The interval to fit. If None, these default to x[0] and x[-1]
respectively.
k : int, optional
The degree of the spline fit. It is recommended to use cubic splines.
Even values of k should be avoided especially with small s values.
1 <= k <= 5
task : {1, 0, -1}, optional
If task==0 find t and c for a given smoothing factor, s.
If task==1 find t and c for another value of the smoothing factor, s.
There must have been a previous call with task=0 or task=1 for the same
set of data (t will be stored an used internally)
If task=-1 find the weighted least square spline for a given set of
knots, t. These should be interior knots as knots on the ends will be
added automatically.
s : float, optional
A smoothing condition. The amount of smoothness is determined by
satisfying the conditions: sum((w * (y - g))**2,axis=0) <= s where g(x)
is the smoothed interpolation of (x,y). The user can use s to control
the tradeoff between closeness and smoothness of fit. Larger s means
more smoothing while smaller values of s indicate less smoothing.
Recommended values of s depend on the weights, w. If the weights
represent the inverse of the standard-deviation of y, then a good s
value should be found in the range (m-sqrt(2*m),m+sqrt(2*m)) where m is
the number of datapoints in x, y, and w. default : s=m-sqrt(2*m) if
weights are supplied. s = 0.0 (interpolating) if no weights are
supplied.
t : array_like, optional
The knots needed for task=-1. If given then task is automatically set
to -1.
full_output : bool, optional
If non-zero, then return optional outputs.
per : bool, optional
If non-zero, data points are considered periodic with period x[m-1] -
x[0] and a smooth periodic spline approximation is returned. Values of
y[m-1] and w[m-1] are not used.
quiet : bool, optional
Non-zero to suppress messages.
This parameter is deprecated; use standard Python warning filters
instead.
Returns
-------
tck : tuple
A tuple (t,c,k) containing the vector of knots, the B-spline
coefficients, and the degree of the spline.
fp : array, optional
The weighted sum of squared residuals of the spline approximation.
ier : int, optional
An integer flag about splrep success. Success is indicated if ier<=0.
If ier in [1,2,3] an error occurred but was not raised. Otherwise an
error is raised.
msg : str, optional
A message corresponding to the integer flag, ier.
See Also
--------
UnivariateSpline, BivariateSpline
splprep, splev, sproot, spalde, splint
bisplrep, bisplev
BSpline
make_interp_spline
Notes
-----
See `splev` for evaluation of the spline and its derivatives. Uses the
FORTRAN routine ``curfit`` from FITPACK.
The user is responsible for assuring that the values of `x` are unique.
Otherwise, `splrep` will not return sensible results.
If provided, knots `t` must satisfy the Schoenberg-Whitney conditions,
i.e., there must be a subset of data points ``x[j]`` such that
``t[j] < x[j] < t[j+k+1]``, for ``j=0, 1,...,n-k-2``.
This routine zero-pads the coefficients array ``c`` to have the same length
as the array of knots ``t`` (the trailing ``k + 1`` coefficients are ignored
by the evaluation routines, `splev` and `BSpline`.) This is in contrast with
`splprep`, which does not zero-pad the coefficients.
References
----------
Based on algorithms described in [1]_, [2]_, [3]_, and [4]_:
.. [1] P. Dierckx, "An algorithm for smoothing, differentiation and
integration of experimental data using spline functions",
J.Comp.Appl.Maths 1 (1975) 165-184.
.. [2] P. Dierckx, "A fast algorithm for smoothing data on a rectangular
grid while using spline functions", SIAM J.Numer.Anal. 19 (1982)
1286-1304.
.. [3] P. Dierckx, "An improved algorithm for curve fitting with spline
functions", report tw54, Dept. Computer Science,K.U. Leuven, 1981.
.. [4] P. Dierckx, "Curve and surface fitting with splines", Monographs on
Numerical Analysis, Oxford University Press, 1993.
Examples
--------
>>> import matplotlib.pyplot as plt
>>> from scipy.interpolate import splev, splrep
>>> x = np.linspace(0, 10, 10)
>>> y = np.sin(x)
>>> spl = splrep(x, y)
>>> x2 = np.linspace(0, 10, 200)
>>> y2 = splev(x2, spl)
>>> plt.plot(x, y, 'o', x2, y2)
>>> plt.show()
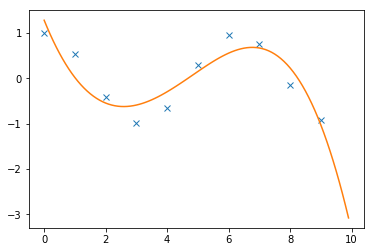
Interpolate between points:
In [23]:
x = np.arange(10)
y = np.cos(x)
Create interpolator
In [24]:
ay = interpolate.splrep(x,y,s=1)
Interpolate
In [25]:
xi = np.arange(0,10,0.1)
yi = interpolate.splev(xi,ay)
Plot:
In [26]:
import pylab as plt
plt.plot(x,y,'x',xi,yi)
plt.show()
In [ ]:
