Modeling a Gaussian Process over heterogeneous time series#
We consider here a physical process, such as stellar activity modulated by stellar rotation, affecting several observables (3 here) differently (different amplitudes, lags, etc.). We will model this physical process through a Gaussian process and its derivative. In this model, each observable is a linear combination of the GP (\(G(t)\)) and its derivative (\(G'(t)\)):
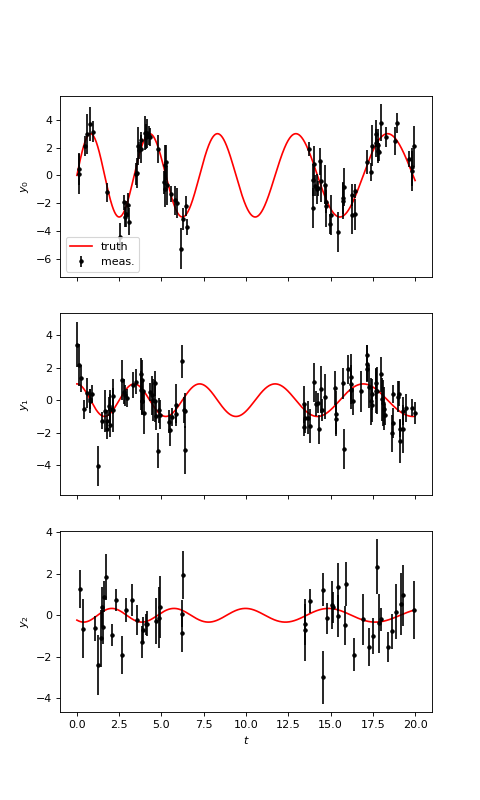
We first generate the 3 time series with a quasiperiodic evolution and different lags:
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(0)
# Settings
P0 = 3.8
dP = 1.25
tmax = 20
amp = [3.0, 1.0, 0.33]
phase = [0, np.pi / 2, -3*np.pi / 4]
nt = [75, 100, 50]
# True signal
tsmooth = np.linspace(0, tmax, 2000)
Psmooth = P0 + dP * (tsmooth / tmax - 1 / 2)
Ysignal = [
ak * np.sin(2 * np.pi * tsmooth / Psmooth + pk)
for ak, pk in zip(amp, phase)
]
# Generate observations calendars
T = [
np.sort(
np.concatenate((np.random.uniform(0, tmax / 3,
ntk // 2), np.random.uniform(2 * tmax / 3, tmax, (ntk + 1) // 2))))
for ntk in nt
]
# Generate measurements with white noise
Yerr = [np.random.uniform(0.5, 1.5, ntk) for ntk in nt]
P = [P0 + dP * (tk / tmax - 1 / 2) for tk in T]
Y = [
amp[k] * np.sin(2 * np.pi * T[k] / P[k] + phase[k]) +
np.random.normal(0, Yerr[k]) for k in range(3)
]
# Plot
_, axs = plt.subplots(3, 1, sharex=True, figsize=(6, 10))
for k in range(3):
ax = axs[k]
ax.plot(tsmooth, Ysignal[k], 'r', label='truth')
ax.errorbar(T[k], Y[k], Yerr[k], fmt='.', color='k', label='meas.')
ax.set_ylabel(f'$y_{k}$')
ax.set_xlabel('$t$')
axs[0].legend()
(Source code, png, hires.png, pdf)
We now fit these data using S+LEAF:
from spleaf import cov, term
from scipy.optimize import fmin_l_bfgs_b
# Merge all 3 time series
t_full, y_full, yerr_full, series_index = cov.merge_series(T, Y, Yerr)
# Initialize the S+LEAF model
C = cov.Cov(t_full,
err=term.Error(yerr_full),
GP=term.MultiSeriesKernel(term.SHOKernel(1.0, 5.0, 1.0), series_index,
np.ones(3), np.ones(3)))
# Fit the hyperparameters using the fmin_l_bfgs_b function from scipy.optimize.
# List of parameters to fit
param = C.param[1:]
# The amplitude of the SHOKernel is fixed at 1 (not fitted),
# since it would be degenerated with the amplitudes alpha, \beta.
# Define the function to minimize
def negloglike(x, y, C):
C.set_param(x, param)
nll = -C.loglike(y)
# gradient
nll_grad = -C.loglike_grad()[1][1:]
return (nll, nll_grad)
# Fit
xbest, _, _ = fmin_l_bfgs_b(negloglike, C.get_param(param), args=(y_full, C))
# Use S+LEAF to predict the missing data
C.set_param(xbest, param)
_, axs = plt.subplots(3, 1, sharex=True, figsize=(6, 10))
for k in range(3):
# Predict time series k
C.kernel['GP'].set_conditional_coef(series_id=k)
mu, var = C.conditional(y_full, tsmooth, calc_cov='diag')
# Plot
ax = axs[k]
ax.plot(tsmooth, Ysignal[k], 'r', label='truth')
ax.errorbar(T[k], Y[k], Yerr[k], fmt='.', color='k', label='meas.')
ax.fill_between(tsmooth,
mu - np.sqrt(var),
mu + np.sqrt(var),
color='g',
alpha=0.5)
ax.plot(tsmooth, mu, 'g', label='predict.')
ax.set_ylabel(f'$y_{k}$')
ax.set_xlabel('$t$')
axs[0].legend()
plt.show()
(Source code, png, hires.png, pdf)
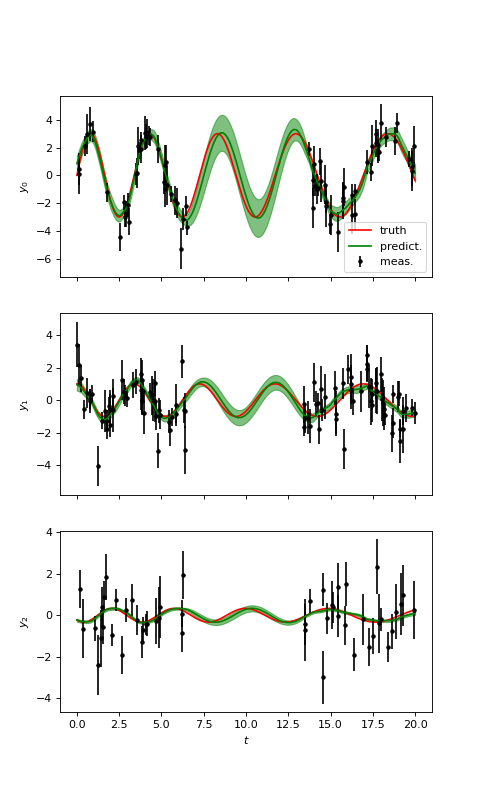
Thanks to the informations contained in the two first time series \(y_0\) and \(y_1\), we obtain a good prediction for the third time series \(y_2\), even if the signal to noise ratio is low.